ATLASea is one of the laureates of the PEPR exploratory programmes, which target emerging scientific or technological sectors where the French government sees a need to identify and structure communities. Co-led by the CNRS and the CEA, the ATLASea programme is funded by France 2030 over 8 years. Its aim is to sequence the genomes of 4,500 eukaryotic marine species (species whose cells have a nucleus), including molluscs, crustaceans, annelids, cnidarians, ascidians, unicellular and multicellular algae, sponges and fish, i.e. around a third of known marine species in France and the overseas territories. The data collected will be deposited in an open-access database for the scientific community, thus completing biodiversity inventories. Under the aegis of the Earth Biogenome Project (EBP), there are more than 50 international programmes aimed at obtaining reference genomes.
Why sequence the genomes of marine species
The phenomenal advances in sequencing technologies over the last two decades are opening up unprecedented possibilities for understanding living organisms. There are an estimated 8.7 million eukaryotic species in the world, including around 2.2 million marine species. Efforts to target microfauna and molecular analysis could increase the number of species identified, especially as the marine world has the largest number of exclusive lineages, i.e. organisms that have never left the ocean. This diversity of species is reflected in the remarkable and largely understudied ways in which they exist and adapt to their environment. This is why ATLASea aims to produce a reference genomic data base on marine organisms in the French exclusive economic zone.
Organisms are linked by their DNA, inherited from common ancestors since the origin of life. For each organism, this genome contains all the instructions necessary for the development of the organism and the functioning of the adult. This data will enable us to understand the mechanisms underlying the physiology of these species, and to retrace their evolutionary history through their genomes. Indeed, analysing the genomes of modern species is an effective way of going back in time, and gaining access to genetic information about their long-lost ancestors. The sequencing of thousands of eukaryotic species is therefore a powerful tool for tracing this history and gaining a better understanding of the origins of life. In addition, a better understanding of marine biodiversity and its dynamics will help to better preserve marine environments. Finally, the discovery of new molecules of interest will be useful for health, agriculture and food.
Stages of the ATLASea project
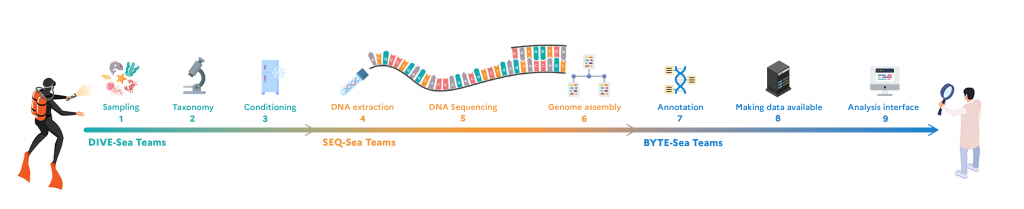
To obtain the expected results, fishermen, divers and researchers will first have to collect marine species at around ten sites spread along the French coastline and overseas (New Caledonia, Guadeloupe, Mayotte, French Polynesia), covering a range of climatic, geomorphological and environmental conditions. A wide range of collection methodologies will be deployed to maximise the diversity of species and taxonomic groups sampled: boats will deploy sampling equipment of different sizes and adapted to different depths or types of seabed. Divers will be using screw-capped vials for visual sampling, marine hoovers for sampling from the surface of rocks and soft bottoms, and baskets and brushes for collecting fauna hiding under rocks. Marine mammal tissues will only be collected from naturally occurring strandings. At the same time, species will be cultivated at the marine stations in Roscoff, Banyuls and Villefranche-sur-Mer.
Once collected, the specimens are identified and classified by experienced taxonomists at the Muséum national d’Histoire naturelle (MNHN), who will check their sequencing status in international databases. If they have not already been declared there, the next step is to quickly immerse the specimens in liquid nitrogen at -196°C, so that they freeze instantly, then preserve them in a thick polystyrene box filled with dry ice at -80°C.
Now it’s off to the Genoscope – National Sequencing Centre. During the transfer, the cold chain must be rigorously observed, as freezing the tissues prevents the DNA from degrading. The CEA’s biology engineers then retrieve the samples and can begin extracting the DNA from the tissues. This is an arduous task, as the DNA of certain organisms such as algae is difficult to extract. New purification protocols have to be put in place to obtain high concentrations of DNA. Before embarking on the complete sequencing of the genome, ‘light’ sequencing is carried out to confirm the identification of the species, especially if identification on the basis of the morphology of the specimen alone was not sufficient. Next comes the genome sequencing stage, using different techniques depending on the information required (Illumina, Pacific Bioscience, Oxford Nanopore Technologies). Sequencing techniques are not capable of sequencing entire chromosomes; they must first fragment them into larger or smaller pieces. Fortunately, however, the latest-generation methods are capable of sequencing very long fragments at high throughput, which can then be reassembled to form a whole genome sequence.
Once assembled, the genome is ready to be analysed: this is the annotation phase, which consists of extracting characteristics such as the location of genes, their structure, size and functions, using computer tools. To do this, the bioinformaticians in charge will use automated analysis methods to annotate the genomes more quickly. All the information from the genomes will be stored at the Institut Français de Bioinformatique (IFB) and will be accessible via a portal that offer visualisation tools for comparative genomics and the reconstruction of ancestral genomes. At the same time, the MNHN will store the reference specimens, from which the tissues have been taken, at -80˚C, thus constituting naturalist collections of the future.
Our commitments: environment and open science
Sampling campaigns are designed to leave the smallest possible footprint on the environments visited. In addition, ATLASea complies with current regulations on protected species and areas, conventional collection methods and the use of animals for scientific purposes. Scientific practice in terms of data management is part of a FAIR approach (easy to Find, Accessible, Interoperable and Reusable). Cryopreservation having also its carbon footprint, the number of samples stored at -80°C will be limited to the strict necessary. Finally, ATLASea will comply with the Nagoya protocol on access to genetic resources and the fair and equitable sharing of benefits arising from their use.
Regulatory Compliance, Ethics & Best Practice Statement
Prior to any submission of samples for sequencing and its metadata in the framework of ATLASea programme, partners of the ATLASea programme (see institutional committee in Consortium/Governance) will comply with all applicable laws, licences, permissions, permits, and regulations (both domestic and international in scope) relating to the acquisition, transfer, storage, use, destruction, and disposal (where relevant) of the samples or specimens they collect. They will provide documentary evidence of this compliance on request to the ATLASea Regulatory Compliance Committee. Scientists who are not employed by the ATLAsea programme and who wish to provide samples shall inform their legal representatives who should certify that the requirements of this statement will be respected, and in particular the above.
For all of these activities, ATLASea partners and scientists employed by non-partner institutions must implement ATLASea Standard Operating Procedures (SOPs), which should include details of regulatory and ethical requirements. The standards within this document must be maintained during transfer of samples or specimens from/to ATLASea team partners and any other collaborators.
Feel free to read the full text of the “ATLASea Regulatory Compliance, Ethics & Best Practice Statement” here:
